Autism is strongly associated with 31 gut microbiota (bacteria, archaea, fungi, and viruses)
Multikingdom and functional gut microbiota markers for autism spectrum disorder
Nature Microbiology (2024) https://doi.org/10.1038/s41564-024-01739-1 can rent PDF at DeepDyve
Qi Su, Oscar W. H. Wong, Wenqi Lu, Yating Wan, Lin Zhang, Wenye Xu, Moses K. T. Li, Chengyu Liu, Chun Pan Cheung, Jessica Y. L. Ching, Pui Kuan Cheong, Ting Fan Leung, Sandra Chan, Patrick Leung, Francis K. L. Chan & Siew C. Ng
Associations between the gut microbiome and autism spectrum disorder (ASD) have been investigated although most studies have focused on the bacterial component of the microbiome. Whether gut archaea, fungi and viruses, or function of the gut microbiome, is altered in ASD is unclear. Here we performed metagenomic sequencing on faecal samples from 1,627 children (aged 1–13 years, 24.4% female) with or without ASD, with extensive phenotype data. Integrated analyses revealed that
51 bacteria,
14 archaea,
7 fungi,
18 viruses,
27 microbial genes and
12 metabolic pathways
were altered in children with ASD.
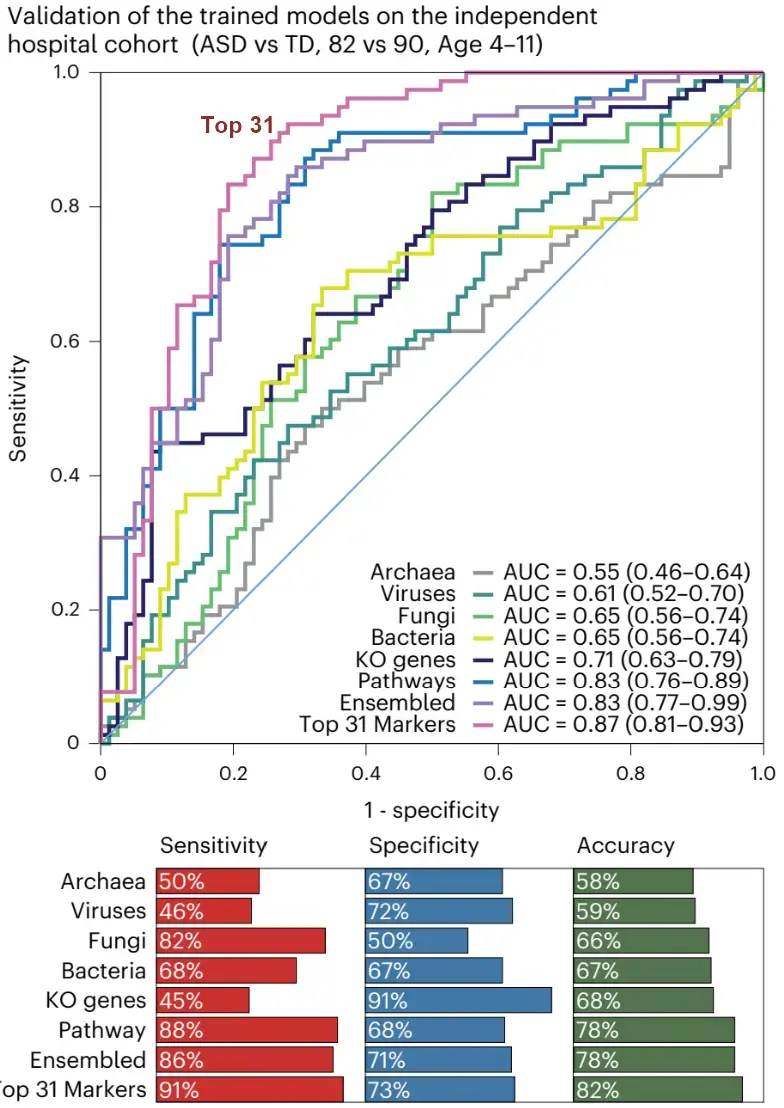
Machine learning using single-kingdom panels showed area under the curve (AUC) of 0.68 to 0.87 in differentiating children with ASD from those that are neurotypical. A panel of 31 multikingdom and functional markers showed a superior diagnostic accuracy with an AUC of 0.91 , with comparable performance for males and females.
Accuracy of the model was predominantly driven by the biosynthesis pathways of ubiquinol-7 or thiamine diphosphate, which were less abundant in children with ASD. Collectively, our findings highlight the potential application of multikingdom and functional gut microbiota markers as non-invasive diagnostic tools in ASD.
Perplexity AI compares gut mirobiota and gut bacteria
Unknown: which causes which; can the microbiota be changed by Vitamin D, etc.
17 Autism risk factors: low Vitamin D, virus, vaccine, mercury etc. - many studies
VitaminDWiki – Microbiome contains
{include}
